Publish Tab
On the "Publish" step, you will generate the files needed for publication or use with downsteam tools. GenSAS outputs the data in GFF3 and FASTA file formats. Any manual curation changes made in the "User-created Annotations" track in JBrowse/Apollo will be merged into the OGS at this step. GenSAS will also add assembly and annotation version numbers to the output files. You can choose which data is exported during this process, but GenSAS automatically selects the minimum files needed (Fig. 64). GenSAS will automatically publish the OGS, the associated gene models and sequences, and the masked repeat consensus. You can select to publish the results from the repeat, gene model prediction, alignment, other structural feature, and functional tools.
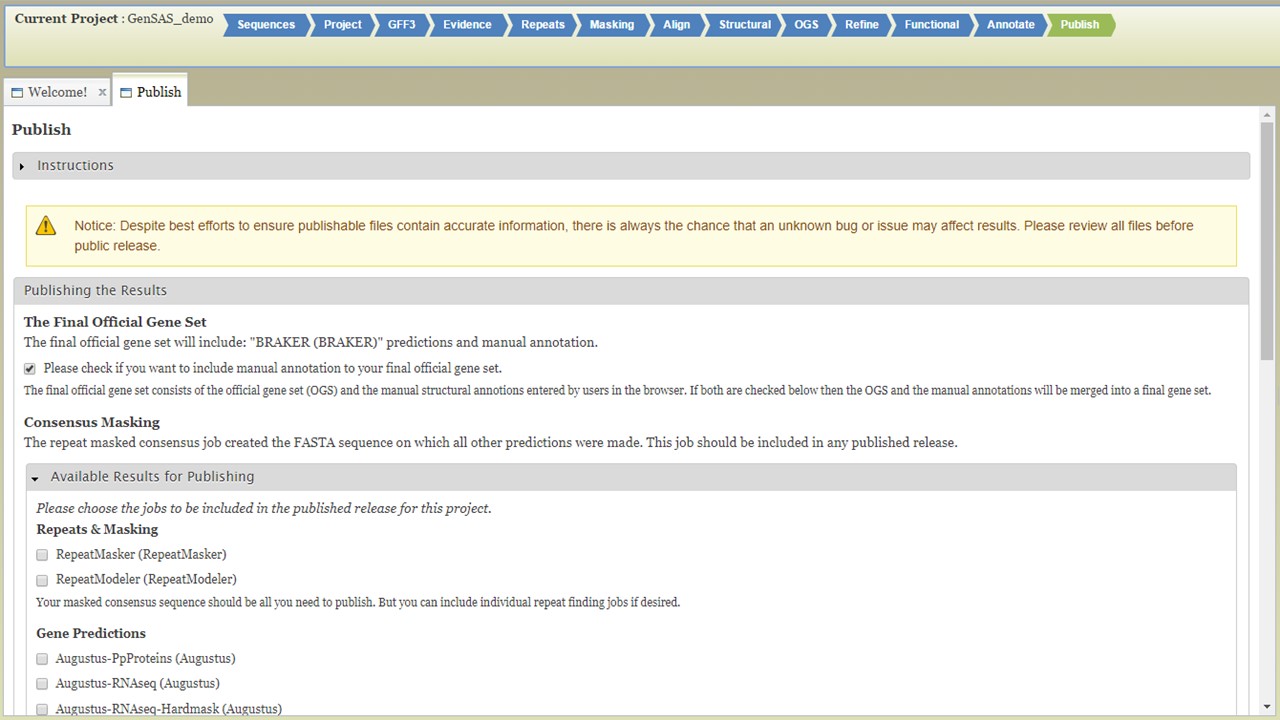
Figure 64. GenSAS "Publish" tab.
Once you have selected the datasets that you want output, click on the "Publish" button and an "Annotations" job will appear in the job queue. If you come back to GenSAS and make changes, you can generate version 2 of the annotation, with the new changes, by running another job under the "Publish" tab. Once the publish job is completed, access and download the files by clicking on the annotation job name in the job queue. Please note that GenSAS will remove proteins under 30 amino acids in length in the final published files. This will open the job summary tab (Fig. 65). The job summary tab has many different sections. At the very top is a button to create a merged GFF3 file (see more below). Nest, is a job summary table which lists which jobs were included in the final files, which track was the OGS, and a link to download a Project Summary file (Fig. 65B) that contains all the settings from the tools used to create the OGS. Below the project summary, is the list of generated files that can be downloaded (Fig. 65C). You can then click on the file names to download the files to your computer. If you want to download all the files at once, click on the "Download all" button at the end of the list (Fig. 65D). At the very bottom of the page is the option to run BUSCO on the predicted proteins of the final annotation (Fig. 65E).
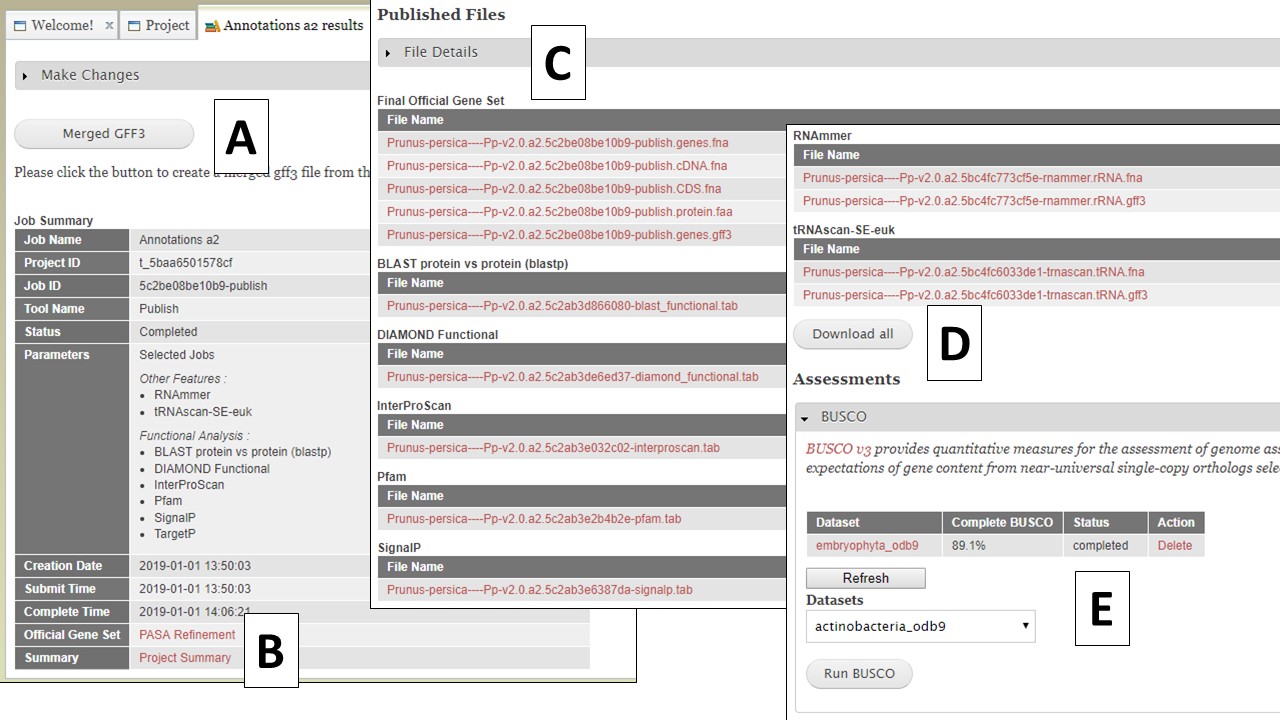
Figure 65. Annotation job results.
The option to create a merged GFF file (Fig. 65A) allows the user to add the repeats from RepeatMasker and Repeat Modeler and the rRNAs and tRNAs from RNAmmer and tRNAScan-SE into the GFF3 file of the OGS (FIg. 66A). The option to also add some of the functional annotation data into column 9 of the GFF3 is also present. After the file is generated, it can be downloaded by opening the "Merged GFF3" interface and clicking on the file name in the table at the very top of the page (Fig. 66B).
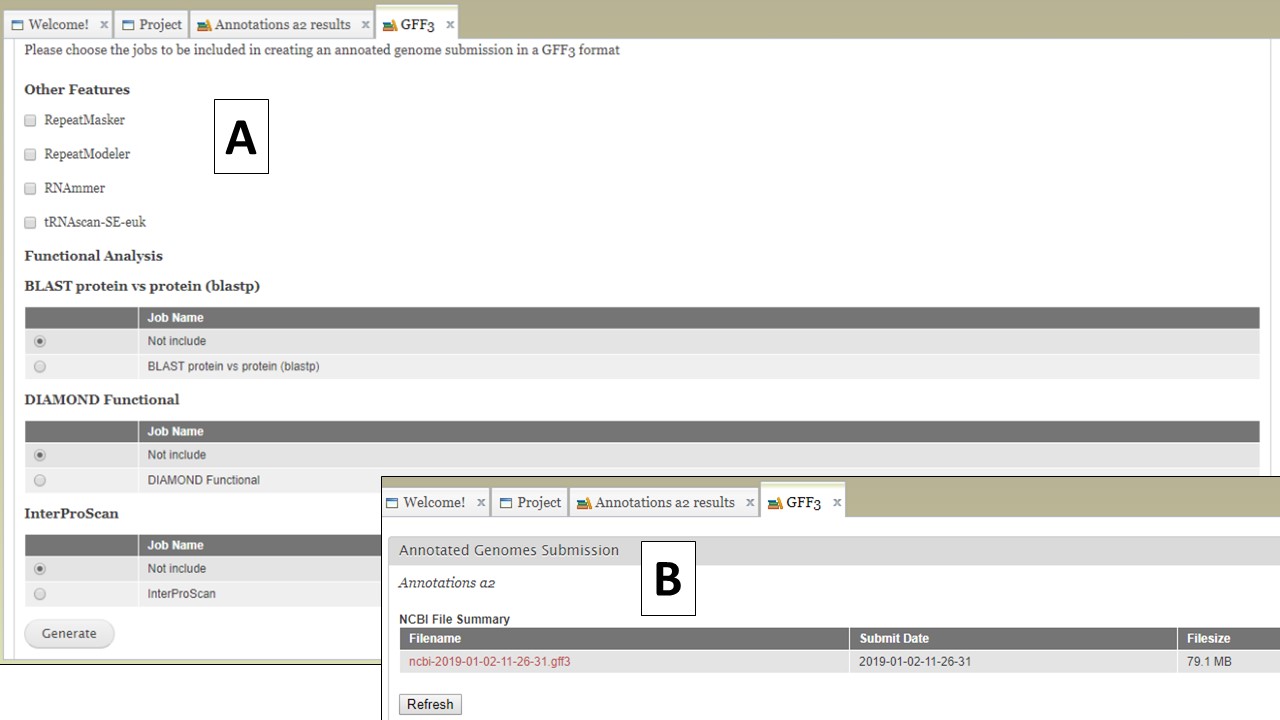
Figure 66. Creating a merged GFF3.




