Evidence Tab
Under the Evidence tab, you can upload evidence files that will be accessible to several tools during the annotation process. Please use data from your species for the best results, but if data from your species is not available, data from a very closely-related species can be used. There are five different evidence types that can be loaded into GenSAS (Fig. 14A). The interface to upload each data type is accessed by clicking on the name (see more details below). The Evidence tab defaults to the "Evidence Files" section that has a table of all evidence files that are associated with your user account (Fig. 14B). If you have uploaded files under another project, they will be available to use in all projects associated with your GenSAS account. As files are uploaded using this tab, you will have to click "Refresh table" (Fig. 14C) to see the files. Files only appear on this table when the upload to GenSAS is complete. Please do not close your browser or log out of GenSAS until the file upload(s) is complete. Large files (i.e. RNA-seq reads) will take a long time to upload.
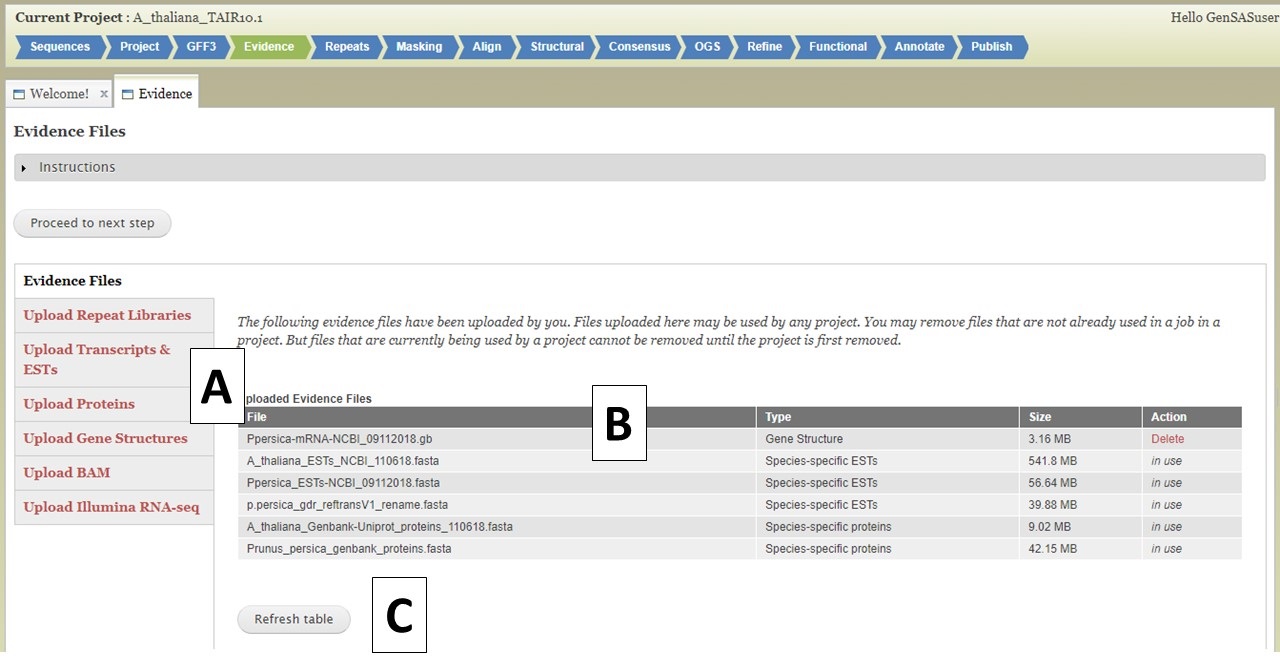
Figure 14. Evidence tab in GenSAS
For the different evidence categories, different file types are accepted. For Repeat Libraries, Transcripts & ESTs, and Proteins, FASTA file formats are accepted. Please see more about the FASTA format under the Sequences section of the User Guide. For the Gene Structures category, a Genbank (.gb) file format is accepted and can be used to train Augustus. For the Genbank gene file, there must be at least 100 sequences in the file for Augustus to use the file during training. BAM files of aligned RNA-seq reads can also be uploaded and used to train Augustus and BRAKER. To add files, click on the appropriate category (Fig. 15A), click on the "Choose File" button to select the file and then click "Upload Files" (Fig. 15B) to start the upload process. You will then see a progress bar appear on the table (Fig. 15C). Do not close your browser or logout of GenSAS until the process has completed. If you close your browser, then the file upload process stops. Once the file has uploaded, you can view the file on the table in the "Evidence Files" section by clicking on "Refresh table" (Fig. 14C).
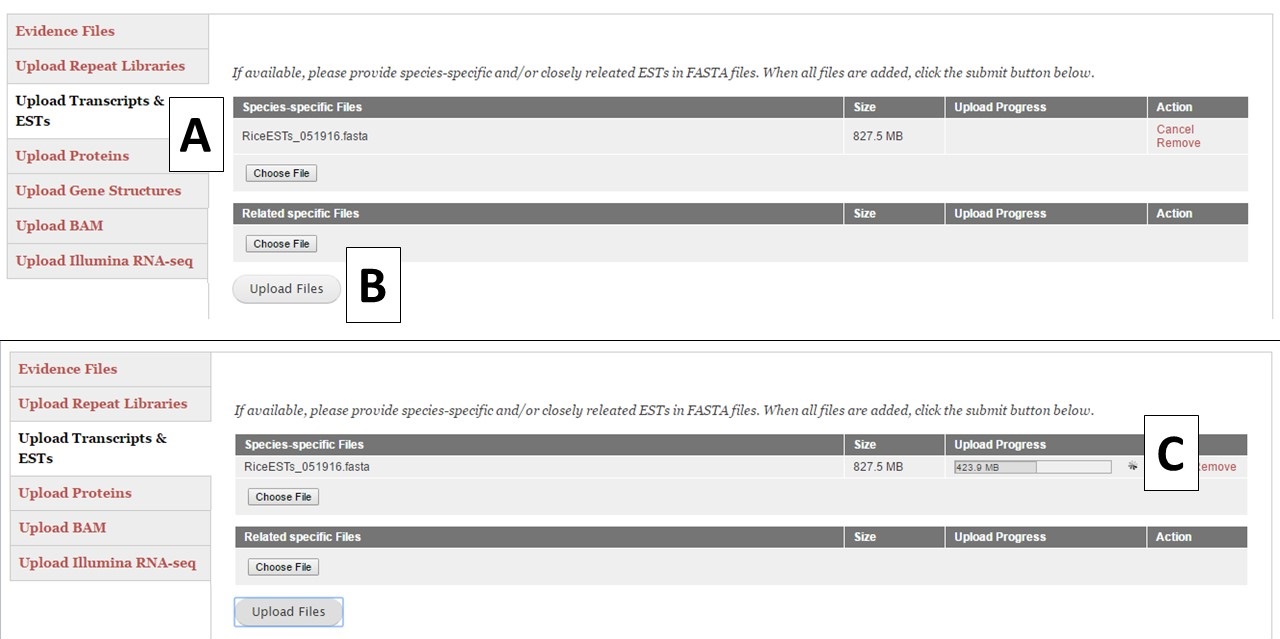
Figure 15. Example file loading process.
Under the "Upload Illumina RNA-seq" section you can upload pre-processed RNA-seq reads from the Illumina platform (Fig. 16). GenSAS currently only supports reads from Illumina. We also ask that the reads have been at least filtered to remove any low-quality reads. You are allowed to load up to 100 GB of RNA-seq data either as a single set of paired reads (Fig. 16A) or one non-paired reads file (Fig. 16B). Please be aware that the upload process may take a significant amount of time due to the file sizes. You may want to consider only loading a portion of your RNA-seq reads into GenSAS. Please note that GenSAS is not a tool for analyzing RNA-seq data. If you are having problems with getting your RNA-seq data uploaded (files > 2 Gb in size) to GenSAS, please contact us. The RNA-seq reads can be aligned to your genome with TopHat and the resulting alignment can be used to train gene model prediction programs. You can also upload assembled RNA-seq data as a FASTA file under the "Upload Transcripts & ESTs" section to use with the other alignment tools in GenSAS.
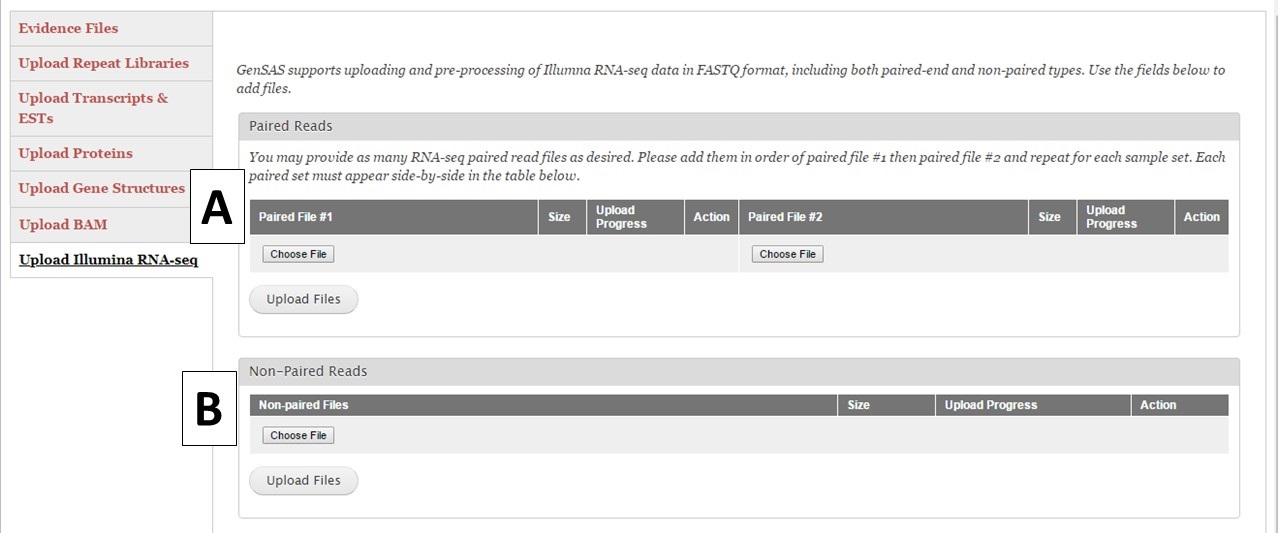
Figure 16. RNA-Seq read upload interface.
When you are done uploading evidence files, click "Proceed to next step", which is located near the top of the tab, to move on to the next step of the annotation process.




